- Record: found
- Abstract: found
- Article: found
A cell-type-specific atlas of the inner ear transcriptional response to acoustic trauma
Read this article at
SUMMARY
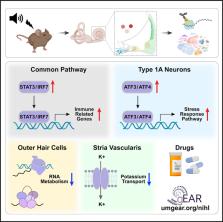
Noise-induced hearing loss (NIHL) results from a complex interplay of damage to the sensory cells of the inner ear, dysfunction of its lateral wall, axonal retraction of type 1C spiral ganglion neurons, and activation of the immune response. We use RiboTag and single-cell RNA sequencing to survey the cell-type-specific molecular landscape of the mouse inner ear before and after noise trauma. We identify induction of the transcription factors STAT3 and IRF7 and immune-related genes across all cell-types. Yet, cell-type-specific transcriptomic changes dominate the response. The ATF3/ATF4 stress-response pathway is robustly induced in the type 1A noise-resilient neurons, potassium transport genes are downregulated in the lateral wall, mRNA metabolism genes are downregulated in outer hair cells, and deafness-associated genes are downregulated in most cell types. This transcriptomic resource is available via the Gene Expression Analysis Resource (gEAR; https://umgear.org/NIHL) and provides a blueprint for the rational development of drugs to prevent and treat NIHL.
In brief
Milon et al. show that cell-type-specific transcriptomic changes following noise exposure dominate the response compared to common changes. The noise-resilient type 1A neurons induce the ATF3/ATF4 stress-response pathway, and the outer hair cells and lateral wall downregulate mRNA metabolism genes and potassium transport genes, respectively.
Graphical Abstract

Related collections
Most cited references111

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.
- Record: found
- Abstract: found
- Article: not found