- Record: found
- Abstract: found
- Article: found
FOXM1c promotes oesophageal cancer metastasis by transcriptionally regulating IRF1 expression
Read this article at
Abstract
Objectives
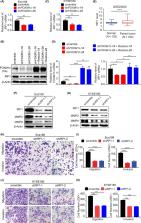
We aimed to elucidate the role and molecular mechanisms of FOXM1 in regulating metastasis in oesophageal squamous cell carcinoma (ESCC) as well as its clinical implications.
Materials and methods
The expression levels of four isoforms of FOXM1 were analysed by real‐time PCR. Next, genetically modification using overexpression and RNAi systems and transwell were employed to examine FOXM1c function in invasion and migration. Dual luciferase and ChIP assays were performed to decipher the underlying mechanism for transcriptional regulation. The expression levels of FOXM1 and IRF1 were determined by immunohistochemistry staining in ESCC specimens.
Results
The FOXM1c was predominantly overexpressed in ESCC cell lines compared to the other FOXM1 isoforms. Ectopic expression of FOXM1c promoted invasion and migration of ESCC cells lines, whereas downregulation of FOXM1c inhibited these processes. Moreover, FOXM1c expression was positively correlated with IRF1 expression in ESCC cell lines and tumour specimens. IRF1 is, at least in part, responsible for FOXM1c‐mediated invasion and migration. Mechanistically, we identified IRF1 as a transcriptional target of FOXM1c and found a FOXM1c‐binding site in the IRF1 promoter region. Furthermore, high expression levels of both FOXM1c and IRF1 were positively associated with low survival rate and predicted a poor prognosis of oesophageal cancer patients.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase.
- Record: found
- Abstract: found
- Article: not found
FOXM1: From cancer initiation to progression and treatment.
- Record: found
- Abstract: found
- Article: not found