- Record: found
- Abstract: found
- Article: found
Error-based or target-based? A unified framework for learning in recurrent spiking networks
Read this article at
Abstract
The field of recurrent neural networks is over-populated by a variety of proposed
learning rules and protocols. The scope of this work is to define a generalized framework,
to move a step forward towards the unification of this fragmented scenario. In the
field of supervised learning, two opposite approaches stand out, error-based and target-based.
This duality gave rise to a scientific debate on which learning framework is the most
likely to be implemented in biological networks of neurons. Moreover, the existence
of spikes raises the question of whether the coding of information is rate-based or
spike-based. To face these questions, we proposed a learning model with two main parameters,
the rank of the feedback learning matrix
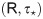

Author summary
Learning in biological or artificial networks means changing the laws governing the
network dynamics in order to better behave in a specific situation. However, there
exists no consensus on what rules regulate learning in biological systems. To face
these questions, we propose a novel theoretical formulation for learning with two
main parameters, the number of learning constraints (


Our approach naturally lends itself to Imitation Learning (and Behavioral Cloning
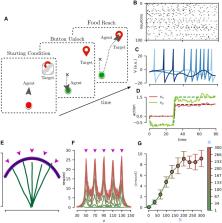
in particular) and we apply it to solve relevant closed-loop tasks such as the button-and-food
task, and the 2D Bipedal Walker. The button-and-food is a navigation task that requires
retaining a long-term memory, and benefits from a high

Finally, we show that our theoretical formulation suggests protocols to deduce the structure of learning feedback in biological networks.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
A cellular mechanism for cortical associations: an organizing principle for the cerebral cortex.

- Record: found
- Abstract: found
- Article: found