- Record: found
- Abstract: found
- Article: found
Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the variant of concern lineage B.1.1.7

Read this article at
- oa journal (via doaj)
- oa repository (via OAI-PMH doi match)
- oa repository (via OAI-PMH doi match)
- oa repository (via OAI-PMH doi match)
- oa repository (via OAI-PMH doi match)
- oa repository (via OAI-PMH doi match)
- oa repository (via OAI-PMH title and first author match)
- oa repository (via OAI-PMH title and first author match)
- oa repository (via OAI-PMH title and first author match)
- oa repository (via OAI-PMH title and first author match)
- oa repository (via OAI-PMH title and first author match)
- oa repository (via pmcid lookup)
- oa repository (via OAI-PMH doi match)
- oa repository (via OAI-PMH doi match)
- oa repository (via OAI-PMH doi match)
Powered by 
Abstract
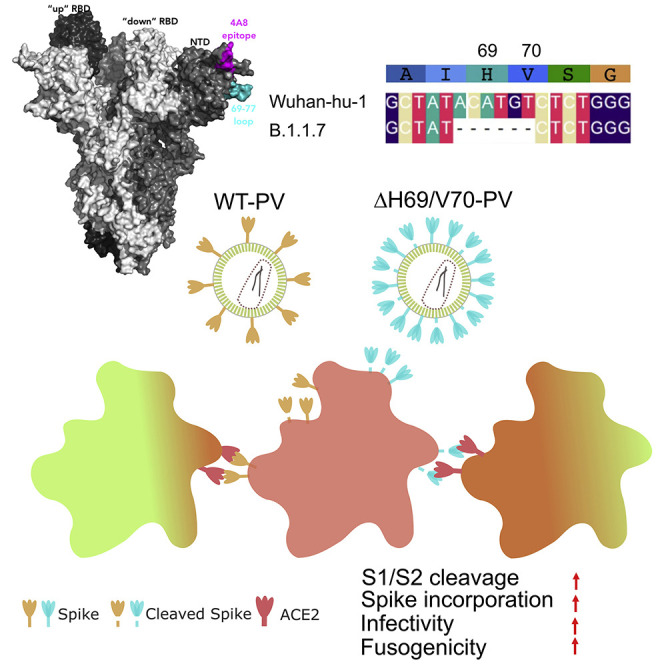
We report SARS-CoV-2 spike ΔH69/V70 in multiple independent lineages, often occurring after acquisition of the receptor binding motif replacements such as N439K and Y453F known to increase binding affinity to the ACE2 receptor and confer antibody escape. In vitro, we show that whilst ΔH69/V70 itself is not an antibody evasion mechanism, it increases infectivity associated with enhanced incorporation of cleaved spike into virions. ΔH69/V70 is able to partially rescue infectivity of S proteins that have acquired N439K and Y453F escape mutations by increased spike incorporation. In addition, replacement of H69 and V70 residues in B.1.1.7 spike (where ΔH69/V70 naturally occurs) impairs spike incorporation and entry efficiency of B.1.1.7 spike pseudotyped virus. B.1.1.7 spike mediates faster kinetics of cell-cell fusion than wild type Wuhan-1 D614G, dependent on ΔH69/V70. Therefore, as ΔH69/V70 compensates for immune escape mutations that impair infectivity, continued surveillance for deletions with functional effects is warranted.
Graphical Abstract
Abstract
Meng et al. report that the SARS-CoV-2 spike ΔH69/V70 deletion has arisen multiple times. The deletion increases entry efficiency, is associated with increased cleaved spike in virions and can compensate for loss of infectivity. The B.1.1.7 spike requires the ΔH69/V70 deletion for efficient cell entry and cell-cell fusion activity.
Related collections
Most cited references74

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found