- Record: found
- Abstract: found
- Article: found
Multiple molecular dynamics simulation of the isoforms of human translation elongation factor 1A reveals reversible fluctuations between "open" and "closed" conformations and suggests specific for eEF1A1 affinity for Ca 2+-calmodulin
Read this article at
Abstract
Background
Eukaryotic translation elongation factor eEF1A directs the correct aminoacyl-tRNA to ribosomal A-site. In addition, eEF1A is involved in carcinogenesis and apoptosis and can interact with large number of non-translational ligands.
There are two isoforms of eEF1A, which are 98% similar. Despite the strong similarity, the isoforms differ in some properties. Importantly, the appearance of eEF1A2 in tissues in which the variant is not normally expressed can be coupled to cancer development.
We reasoned that the background for the functional difference of eEF1A1 and eEF1A2 might lie in changes of dynamics of the isoforms.
Results
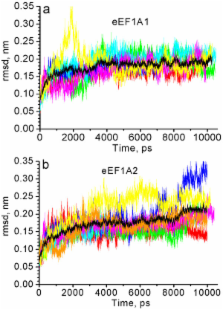
It has been determined by multiple MD simulation that eEF1A1 shows increased reciprocal flexibility of structural domains I and II and less average distance between the domains, while increased non-correlated diffusive atom motions within protein domains characterize eEF1A2. The divergence in the dynamic properties of eEF1A1 and eEF1A2 is caused by interactions of amino acid residues that differ between the two variants with neighboring residues and water environment.
The main correlated motion of both protein isoforms is the change in proximity of domains I and II which can lead to disappearance of the gap between the domains and transition of the protein into a "closed" conformation. Such a transition is reversible and the protein can adopt an "open" conformation again. This finding is in line with our earlier experimental observation that the transition between "open" and "closed" conformations of eEF1A could be essential for binding of tRNA and/or other biological ligands.
The putative calmodulin-binding region Asn311-Gly327 is less flexible in eEF1A1 implying its increased affinity for calmodulin. The ability of eEF1A1 rather than eEF1A2 to interact with Ca2+/calmodulin is shown experimentally in an ELISA-based test.
Conclusion
We have found that reversible transitions between "open" and "close" conformations of eEF1A provide a molecular background for the earlier observation that the eEF1A molecule is able to change the shape upon interaction with tRNA. The ability of eEF1A1 rather than eEF1A2 to interact with calmodulin is predicted by MD analysis and showed experimentally. The differential ability of the eEF1A isoforms to interact with signaling molecules discovered in this study could be associated with cancer-related properties of eEF1A2.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer.
- Record: found
- Abstract: found
- Article: not found
Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization.
- Record: found
- Abstract: not found
- Article: not found