- Record: found
- Abstract: found
- Article: found
Local mitochondrial replication in the periphery of neurons requires the eEF1A1 protein and thetranslation of nuclear-encoded proteins

Read this article at
Summary
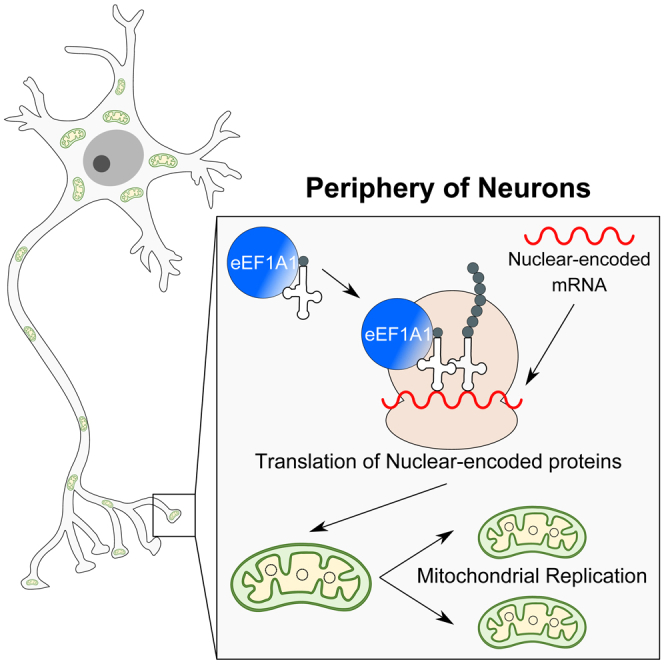
In neurons, it is commonly assumed that mitochondrial replication only occurs in the cell body, after which the mitochondria must travel to the neuron’s periphery. However, while mitochondrial DNA replication has been observed to occur away from the cell body, the specific mechanisms involved remain elusive. Using EdU-labelling in mouse primary neurons, we developed a tool to determine the mitochondrial replication rate. Taking of advantage of microfluidic devices, we confirmed that mitochondrial replication also occurs locally in the periphery of neurons. To achieve this, mitochondria require de novo nuclear-encoded, but not mitochondrial-encoded protein translation. Following a proteomic screen comparing synaptic with non-synaptic mitochondria, we identified two elongation factors – eEF1A1 and TUFM – that were upregulated in synaptic mitochondria. We found that mitochondrial replication is impaired upon the downregulation of eEF1A1, and this is particularly relevant in the periphery of neurons.
Graphical abstract
Highlights
-
•
Mitochondrial replication occurs in distal regions of neurons
-
•
Mitochondrial-encoded translation is not required for mtDNA replication in neurons
-
•
In neurons, mitochondrial replication requires nuclear-encoded protein translation
-
•
EEF1A1 is upregulated at the synapse and is required for mitochondrial replication
Abstract
Biochemistry; Biological sciences; Cellular neuroscience; Molecular neuroscience; Natural sciences; Neuroscience
Related collections
Most cited references60
- Record: found
- Abstract: found
- Article: not found
Universal sample preparation method for proteome analysis.

- Record: found
- Abstract: found
- Article: found
Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0

- Record: found
- Abstract: found
- Article: found