- Record: found
- Abstract: found
- Article: found
Predicting lymph node metastasis from primary tumor histology and clinicopathologic factors in colorectal cancer using deep learning
Read this article at
Abstract
Background
Presence of lymph node metastasis (LNM) influences prognosis and clinical decision-making in colorectal cancer. However, detection of LNM is variable and depends on a number of external factors. Deep learning has shown success in computational pathology, but has struggled to boost performance when combined with known predictors.
Methods
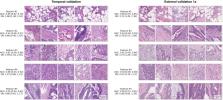
Machine-learned features are created by clustering deep learning embeddings of small patches of tumor in colorectal cancer via k-means, and then selecting the top clusters that add predictive value to a logistic regression model when combined with known baseline clinicopathological variables. We then analyze performance of logistic regression models trained with and without these machine-learned features in combination with the baseline variables.
Results
The machine-learned extracted features provide independent signal for the presence of LNM (AUROC: 0.638, 95% CI: [0.590, 0.683]). Furthermore, the machine-learned features add predictive value to the set of 6 clinicopathologic variables in an external validation set (likelihood ratio test, p < 0.00032; AUROC: 0.740, 95% CI: [0.701, 0.780]). A model incorporating these features can also further risk-stratify patients with and without identified metastasis ( p < 0.001 for both stage II and stage III).
Conclusion
This work demonstrates an effective approach to combine deep learning with established clinicopathologic factors in order to identify independently informative features associated with LNM. Further work building on these specific results may have important impact in prognostication and therapeutic decision making for LNM. Additionally, this general computational approach may prove useful in other contexts.
Plain language summary
When colorectal cancers spread to the lymph nodes, it can indicate a poorer prognosis. However, detecting lymph node metastasis (spread) can be difficult and depends on a number of factors such as how samples are taken and processed. Here, we show that machine learning, which involves computer software learning from patterns in data, can predict lymph node metastasis in patients with colorectal cancer from the microscopic appearance of their primary tumor and the clinical characteristics of the patients. We also show that the same approach can predict patient survival. With further work, our approach may help clinicians to inform patients about their prognosis and decide on appropriate treatments.
Abstract
Krogue et al. develop a deep learning approach for the prediction of lymph node metastasis in patients with colorectal cancer. Computationally-extracted histological features of the primary tumor add predictive value to a set of clinicopathologic factors.
Related collections
Most cited references24
- Record: found
- Abstract: not found
- Article: not found
ImageNet Large Scale Visual Recognition Challenge

- Record: found
- Abstract: found
- Article: found
Predicting cancer outcomes from histology and genomics using convolutional networks
- Record: found
- Abstract: not found
- Article: not found