- Record: found
- Abstract: found
- Article: found
Structural dynamics of the transaminase active site revealed by the crystal structure of a co-factor free omega-transaminase from Vibrio fluvialis JS17
Read this article at
Abstract
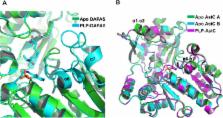
Omega (ω)-transaminase catalyzes the transfer of an amino group from a non-α position amino acid, or an amine compound with no carboxylic group, to an amino acceptor, and has been studied intensively because of its high potential utility in industry and pharmatheutics. The ω-transaminase from Vibrio fluvialis JS17 (Vfat) is an amine:pyruvate transaminase capable of the stereo-selective transamination of arylic chiral amines. This enzyme exhibits extraordinary enantio-selectivity, and has a rapid reaction rate for chiral amine substrates. In this study, we report the crystal structure of the apo form of Vfat. The overall structure of Vfat was typical of other class III aminotransferase exhibiting an N-terminal helical domain, a small domain, and a large domain. Interestingly, the two subunits of apo Vfat in the asymmetric unit had different structures. A comparison of the overall structure to other transaminases, revealed that the structures of the N-terminal helical domain and the large domain can be affected by cofactor occupancy, but the structural rearrangement in these regions can occur independently.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use.
- Record: found
- Abstract: not found
- Article: not found
Dali: a network tool for protein structure comparison.
- Record: found
- Abstract: found
- Article: not found