- Record: found
- Abstract: found
- Article: found
Recent molecular techniques for the diagnosis of Zika and Chikungunya infections: A systematic review
Read this article at
Abstract
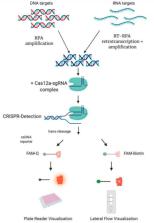
Zika virus (ZIKV) and Chikungunya virus (CHIKV) are arboviruses that cause important viral diseases affecting the world population. Both viruses can produce remarkably similar clinical manifestations, co-circulate in a geographic region, and coinfections have been documented, thus making clinical diagnosis challenging. Therefore, it is urgent to have better molecular techniques that allow a differential, sensitive and rapid diagnosis from body fluid samples. This systematic review explores evidence in the literature regarding the advances in the molecular diagnosis of Zika and Chikungunya in humans, published from 2010 to March 2021. Four databases were consulted (Scopus, PubMed, Web of Science, and Embase) and a total of 31 studies were included according to the selection criteria. Our analysis highlights the need for standardization in the report and interpretation of new promising diagnostic methods. It also examines the benefits of new alternatives for the molecular diagnosis of these arboviruses, in contrast to established methods.
Abstract
Chikungunya, Zika, Nucleic acid amplification techniques, Molecular diagnosis.
Related collections
Most cited references53

- Record: found
- Abstract: found
- Article: found
The PRISMA 2020 statement: an updated guideline for reporting systematic reviews
- Record: found
- Abstract: found
- Article: not found
QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies.
- Record: found
- Abstract: found
- Article: not found