- Record: found
- Abstract: found
- Article: found
The fatal contribution of serine protease-related genetic variants to COVID-19 outcomes
Read this article at
Abstract
Introduction
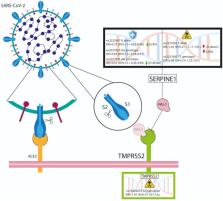
Serine proteases play a critical role during SARS-CoV-2 infection. Therefore, polymorphisms of transmembrane protease serine 2 ( TMPRSS2) and serpine family E member 1 ( SERPINE1) could help to elucidate the contribution of variability to COVID-19 outcomes.
Methods
To evaluate the genetic variants of the genes previously associated with COVID-19 outcomes, we performed a cross-sectional study in which 1536 SARS-CoV-2-positive participants were enrolled. TMPRSS2 (rs2070788, rs75603675, rs12329760) and SERPINE1 (rs2227631, rs2227667, rs2070682, rs2227692) were genotyped using the Open Array Platform. The association of polymorphisms with disease outcomes was determined by logistic regression analysis adjusted for covariates (age, sex, hypertension, type 2 diabetes, and obesity).
Results
According to our codominant model, the GA genotype of rs2227667 (OR=0.55; 95% CI = 0.36-0.84; p=0.006) and the AG genotype of rs2227667 (OR=0.59; 95% CI = 0.38-0.91; p=0.02) of SERPINE1 played a protective role against disease. However, the rs2227692 T allele and TT genotype SERPINE1 (OR=1.45; 95% CI = 1.11-1.91; p=0.006; OR=2.08; 95% CI = 1.22-3.57; p=0.007; respectively) were associated with a decreased risk of death. Similarly, the rs75603675 AA genotype TMPRSS2 had an OR of 1.97 (95% CI = 1.07-3.6; p=0.03) for deceased patients. Finally, the rs2227692 T allele SERPINE1 was associated with increased D-dimer levels (OR=1.24; 95% CI = 1.03-1.48; p=0.02).
Discussion
Our data suggest that the rs75603675 TMPRSS2 and rs2227692 SERPINE1 polymorphisms are associated with a poor outcome. Additionally, rs2227692 SERPINE1 could participate in hypercoagulable conditions in critical COVID-19 patients, and this genetic variant could contribute to the identification of new pharmacological targets and treatment strategies to block the inhibition of TMPRSS2 entry into SARS-CoV-2.
Related collections
Most cited references62

- Record: found
- Abstract: found
- Article: found
A global reference for human genetic variation
- Record: found
- Abstract: found
- Article: not found
Mechanisms of SARS-CoV-2 entry into cells
- Record: found
- Abstract: found
- Article: not found