- Record: found
- Abstract: found
- Article: found
Effective combinatorial immunotherapy for penile squamous cell carcinoma
Read this article at
Abstract
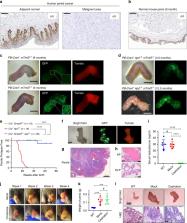
Penile squamous cell carcinoma (PSCC) accounts for over 95% of penile malignancies and causes significant mortality and morbidity in developing countries. Molecular mechanisms and therapies of PSCC are understudied, owing to scarcity of laboratory models. Herein, we describe a genetically engineered mouse model of PSCC, by co-deletion of Smad4 and Apc in the androgen-responsive epithelium of the penis. Mouse PSCC fosters an immunosuppressive microenvironment with myeloid-derived suppressor cells (MDSCs) as a dominant population. Preclinical trials in the model demonstrate synergistic efficacy of immune checkpoint blockade with the MDSC-diminishing drugs cabozantinib or celecoxib. A critical clinical problem of PSCC is chemoresistance to cisplatin, which is induced by Pten deficiency on the backdrop of Smad4/Apc co-deletion. Drug screen studies informed by targeted proteomics identify a few potential therapeutic strategies for PSCC. Our studies have established what we believe to be essential resources for studying PSCC biology and developing therapeutic strategies.
Abstract
Penile squamous cell carcinoma (PSCC) is a cancer that is associated with significant mortality. Here, the authors develop a mouse model of PSCC by co-deletion of Smad4 and Apc in the androgen-responsive penile epithelium, and show synergistic efficacy of checkpoint therapy with cabozantinib or celecoxib in their model.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma.
- Record: found
- Abstract: found
- Article: not found
Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer.
- Record: found
- Abstract: found
- Article: not found