- Record: found
- Abstract: found
- Article: found
The biotechnological potential of marine bacteria in the novel lineage of Pseudomonas pertucinogena

Read this article at
Summary
Marine habitats represent a prolific source for molecules of biotechnological interest. In particular, marine bacteria have attracted attention and were successfully exploited for industrial applications. Recently, a group of Pseudomonas species isolated from extreme habitats or living in association with algae or sponges were clustered in the newly established Pseudomonas pertucinogena lineage. Remarkably for the predominantly terrestrial genus Pseudomonas, more than half (9) of currently 16 species within this lineage were isolated from marine or saline habitats. Unlike other Pseudomonas species, they seem to have in common a highly specialized metabolism. Furthermore, the marine members apparently possess the capacity to produce biomolecules of biotechnological interest (e.g. dehalogenases, polyester hydrolases, transaminases). Here, we summarize the knowledge regarding the enzymatic endowment of the marine Pseudomonas pertucinogena bacteria and report on a genomic analysis focusing on the presence of genes encoding esterases, dehalogenases, transaminases and secondary metabolites including carbon storage compounds.
Abstract
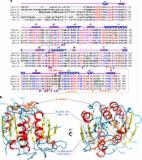
The newly established Pseudomonas pertucinogena lineage consists of several marine bacterial species which possess the capacity to produce biomolecules of biotechnological interest. Beside biocatalysts including dehalogenases, polyester hydrolases, and transaminases they produce several secondary metabolites and carbon storage compounds.
Related collections
Most cited references101

- Record: found
- Abstract: found
- Article: found
antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification
- Record: found
- Abstract: found
- Article: not found
Role of Biocatalysis in Sustainable Chemistry.
- Record: found
- Abstract: found
- Article: found