- Record: found
- Abstract: found
- Article: found
Mouse-Adapted SARS-CoV-2 MA10 Strain Displays Differential Pulmonary Tropism and Accelerated Viral Replication, Neurodissemination, and Pulmonary Host Responses in K18-hACE2 Mice
Read this article at
ABSTRACT
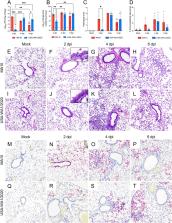
Several models were developed to study the pathogenicity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as well as the in vivo efficacy of vaccines and therapeutics. Since wild-type mice are naturally resistant to infection by ancestral SARS-CoV-2 strains, several transgenic mouse models expressing human angiotensin-converting enzyme 2 (hACE2) were developed. An alternative approach has been to develop mouse-adapted SARS-CoV-2 strains. Here, we compared the clinical progression, viral replication kinetics and dissemination, pulmonary tropism, and host innate immune response dynamics between the mouse-adapted MA10 strain and its parental strain (USA-WA1/2020) following intranasal inoculation of K18-hACE2 mice, a widely used model. Compared to its parental counterpart, the MA10 strain induced earlier clinical decline with significantly higher viral replication and earlier neurodissemination. Importantly, the MA10 strain also showed a wider tropism, with infection of bronchiolar epithelia. While both SARS-CoV-2 strains induced comparable pulmonary cytokine/chemokine responses, many proinflammatory and monocyte-recruitment chemokines, such as interleukin-6 (IL-6), tumor necrosis factor alpha (TNF-α), IP-10/CXCL10, and MCP-1/CCL2, showed an earlier peak in MA10-infected mice. Furthermore, both strains induced a similar downregulation of murine Ace2, with only a transient downregulation of Tmprss2 and no alterations in hACE2 expression. Overall, these data demonstrate that in K18-hACE2 mice, the MA10 strain has a pulmonary tropism that more closely resembles SARS-CoV-2 tropism in humans (airways and pneumocytes) than its parental strain. Its rapid replication and neurodissemination and early host pulmonary responses can have a significant impact on the clinical outcomes of infection and are, therefore, critical features to consider for study designs using these strains and mouse model.
IMPORTANCE The COVID-19 pandemic, caused by SARS-CoV-2, is still significantly impacting health care systems around the globe. Refined animal models are needed to study SARS-CoV-2 pathogenicity as well as efficacy of vaccines and therapeutics. In line with this, thorough evaluation of animal models and virus strains/variants are paramount for standardization and meaningful comparisons. Here, we demonstrated differences in replication dynamics between the Wuhan-like USA-WA1/2020 strain and the derivative mouse-adapted MA10 strain in K18-hACE2 mice. The MA10 strain showed accelerated viral replication and neurodissemination, differential pulmonary tropism, and earlier pulmonary innate immune responses. The observed differences allow us to better refine experimental designs when considering the use of the MA10 strain in the widely utilized K18-hACE2 murine model.
Related collections
Most cited references82

- Record: found
- Abstract: found
- Article: found
A pneumonia outbreak associated with a new coronavirus of probable bat origin
- Record: found
- Abstract: found
- Article: not found
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor

- Record: found
- Abstract: found
- Article: found