- Record: found
- Abstract: found
- Article: found
Clinico-Epidemiological Laboratory Findings of COVID- 19 Positive Patients in a Hospital in Saudi Arabia
Abstract
Background
Understanding COVID-19’s onset and clinical effects requires knowing host immune responses.
Objective
To investigate the presence of IgM, IgG, and cytokine levels (IL-2 and IL-6) in individuals with COVID-19 who have had their diagnosis confirmed by PCR.
Methods
This cross-sectional research included 70 adult ICU patients from King Abdullah Hospital in Bisha, Saudi Arabia. Subjects gave two blood samples. After hospital release, only 21 patients provided the second sample. Each patient provided a sample upon admission. Quantitative ELISAs evaluated IL-2, IL-6, and SARS-CoV-2-specific IgM and IgG antibodies.
Results
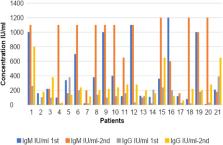
All patients were critically ill and unvaccinated against COVID-19. 46 (65.7%) of the patients were male, and their age range was 33–98 years (with a mean age of 66.5); 24.3%) were 51–61 years old. IgG was positive in all patients, although IgM predominated in 57/70 (81.4%) (6–1200 IU/mL). Total data analysis yielded these results. IL-6 was calculated at 10–1900 ng/mL, whereas IL-2 was 4–280. Discharged hospital patients had a statistically significant increase in IgM and IgG (P = 0.01, 0.004) but a statistically insignificant decline in IL-6 and IL-2 (P = 0.761, 0.071). Low IgM levels increased hospital stays. The study found lengthier hospital stays with higher IgG levels.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China
- Record: found
- Abstract: found
- Article: not found
A Novel Coronavirus from Patients with Pneumonia in China, 2019
- Record: found
- Abstract: found
- Article: not found