- Record: found
- Abstract: found
- Article: found
Systematic evaluation of cell-type deconvolution pipelines for sequencing-based bulk DNA methylomes
Read this article at
Abstract
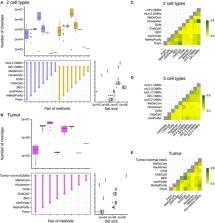
DNA methylation analysis by sequencing is becoming increasingly popular, yielding methylomes at single-base pair and single-molecule resolution. It has tremendous potential for cell-type heterogeneity analysis using intrinsic read-level information. Although diverse deconvolution methods were developed to infer cell-type composition based on bulk sequencing-based methylomes, systematic evaluation has not been performed yet. Here, we thoroughly benchmark six previously published methods: Bayesian epiallele detection, DXM, PRISM, csmFinder+coMethy, ClubCpG and MethylPurify, together with two array-based methods, MeDeCom and Houseman, as a comparison group. Sequencing-based deconvolution methods consist of two main steps, informative region selection and cell-type composition estimation, thus each was individually assessed. With this elaborate evaluation, we aimed to establish which method achieves the highest performance in different scenarios of synthetic bulk samples. We found that cell-type deconvolution performance is influenced by different factors depending on the number of cell types within the mixture. Finally, we propose a best-practice deconvolution strategy for sequencing data and point out limitations that need to be handled. Array-based methods—both reference-based and reference-free—generally outperformed sequencing-based methods, despite the absence of read-level information. This implies that the current sequencing-based methods still struggle with correctly identifying cell-type-specific signals and eliminating confounding methylation patterns, which needs to be handled in future studies.
Related collections
Most cited references60

- Record: found
- Abstract: found
- Article: found
The Sequence Alignment/Map format and SAMtools
- Record: found
- Abstract: not found
- Article: not found
Cutadapt removes adapter sequences from high-throughput sequencing reads

- Record: found
- Abstract: found
- Article: found