- Record: found
- Abstract: found
- Article: found
Abiotic selection of microbial genome size in the global ocean
Read this article at
Abstract
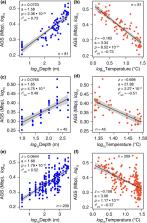
Strong purifying selection is considered a major evolutionary force behind small microbial genomes in the resource-poor photic ocean. However, very little is currently known about how the size of prokaryotic genomes evolves in the global ocean and whether patterns reflect shifts in resource availability in the epipelagic and relatively stable deep-sea environmental conditions. Using 364 marine microbial metagenomes, we investigate how the average genome size of uncultured planktonic prokaryotes varies across the tropical and polar oceans to the hadal realm. We find that genome size is highest in the perennially cold polar ocean, reflecting elongation of coding genes and gene dosage effects due to duplications in the interior ocean microbiome. Moreover, the rate of change in genome size due to temperature is 16-fold higher than with depth up to 200 m. Our results demonstrate how environmental factors can influence marine microbial genome size selection and ecological strategies of the microbiome.
Abstract
This study investigates the average genome size of planktonic prokaryotes across tropical and polar oceans and down to the hadal realm. Using hundreds of metagenomes of marine microorganisms, genome size was found to be highest in the perennially cold polar ocean, suggesting that environmental factors influence genome size selection and the ecological strategies of marine microbes.
Related collections
Most cited references80

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found