- Record: found
- Abstract: found
- Article: found
Graphene BioFET sensors for SARS-CoV-2 detection: a multiscale simulation approach†
Read this article at
Abstract
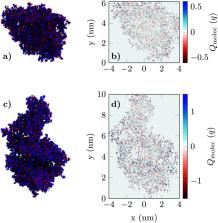
Biological Field-Effect Transistors (BioFETs) have already demonstrated enormous potential for detecting minute amounts of ions and molecules. The use of two-dimensional (2D) materials has been shown to boost their performance and to enable the design of new applications. This combination deserves special interest in the current pandemic caused by the SARS-CoV-2 virus which demands fast, reliable and cheap detection methods. However, in spite of the experimental advances, there is a lack of a comprehensive and in-depth computational approach to capture the mechanisms underlying the sensor behaviour. Here, we present a multiscale platform that combines detailed atomic models of the molecules with mesoscopic device-level simulations. The fine-level description exploited in this approach accounts for the charge distribution of the receptor, its reconfiguration when the target binds to it, and the consequences in terms of sensitivity on the transduction mechanism. The results encourage the further exploration of improved sensor designs and 2D materials combined with diverse receptors selected to achieve the desired specificity.
Abstract
Two-dimensional (2D) materials BioFETs have already demonstrated their potential for detecting low amounts of molecules. Here, we present a multiscale simulation platform in the context of Graphene BioFET for the detection of SARS-CoV-2.

Related collections
Most cited references47

- Record: found
- Abstract: found
- Article: found
Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation
- Record: found
- Abstract: found
- Article: not found
Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein
- Record: found
- Abstract: found
- Article: not found
Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.
