- Record: found
- Abstract: found
- Article: found
A subunit of the oligosaccharyltransferase complex is required for interspecific gametophyte recognition in Arabidopsis
Read this article at
Abstract
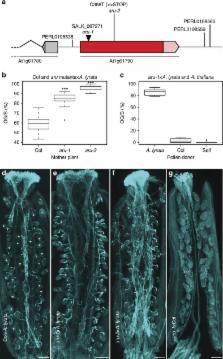
Species-specific gamete recognition is a key premise to ensure reproductive success and the maintenance of species boundaries. During plant pollen tube (PT) reception, gametophyte interactions likely allow the species-specific recognition of signals from the PT (male gametophyte) by the embryo sac (female gametophyte), resulting in PT rupture, sperm release, and double fertilization. This process is impaired in interspecific crosses between Arabidopsis thaliana and related species, leading to PT overgrowth and a failure to deliver the sperm cells. Here we show that ARTUMES ( ARU) specifically regulates the recognition of interspecific PTs in A. thaliana. ARU, identified in a genome-wide association study (GWAS), exclusively influences interspecific—but not intraspecific—gametophyte interactions. ARU encodes the OST3/6 subunit of the oligosaccharyltransferase complex conferring protein N-glycosylation. Our results suggest that glycosylation patterns of cell surface proteins may represent an important mechanism of gametophyte recognition and thus speciation.
Abstract
 Species-specific gamete recognition is needed to maintain species boundaries. Here,
Müller
et al. show that
ARTUMES regulates pollen tube recognition between different
Arabidopsis species, representing the first gene known to exclusively influence inter- but not
intraspecific gamete interaction in plants.
Species-specific gamete recognition is needed to maintain species boundaries. Here,
Müller
et al. show that
ARTUMES regulates pollen tube recognition between different
Arabidopsis species, representing the first gene known to exclusively influence inter- but not
intraspecific gamete interaction in plants.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants.
- Record: found
- Abstract: found
- Article: not found
An efficient multi-locus mixed model approach for genome-wide association studies in structured populations
- Record: found
- Abstract: found
- Article: not found