- Record: found
- Abstract: found
- Article: not found
Size and duration of COVID-19 clusters go along with a high SARS-CoV-2 viral load: A spatio-temporal investigation in Vaud state, Switzerland

Read this article at
Abstract
To understand the geographical and temporal spread of SARS-CoV-2 during the first documented wave of infection in the state of Vaud, Switzerland, we analyzed clusters of positive cases using the precise residential location of 33,651 individuals tested (RT-PCR) between January 10 and June 30, 2020. We used a prospective Poisson space-time scan statistic (SaTScan) and a Modified Space–Time Density-Based Spatial Clustering of Application with Noise (MST-DBSCAN) to identify both space-time and transmission clusters, and estimated cluster duration, transmission behavior (emergence, growth, reduction, etc.) and relative risk. For each cluster, we computed the number of individuals, the median age of individuals and their viral load.
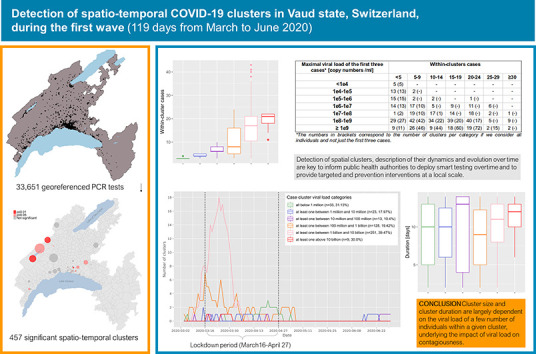
Among the 1684 space-time clusters identified, 457 (27.1%) were significant ( p ≤ 0.05), such that they harbored a higher relative risk of infection within the cluster than compared to regions outside the cluster. Clusters lasted a median of 11 days (IQR 7–13) and included a median of 12 individuals per cluster (IQR 5–20). The majority of significant clusters ( n = 260; 56.9%) had at least one person with an extremely high viral load (>1 billion copies/ml). Those clusters were considerably larger (median of 17 infected individuals, p < 0.001) than clusters with individuals showing a viral load below 1 million copies/ml (median of three infected individuals). The highest viral loads were found in clusters with the lowest average age group considered in the investigation, while clusters with the highest average age had low to middle viral load. In 20 significant clusters, the viral load of the three first cases was below 100,000 copies/ml, suggesting that subjects with fewer than 100,000 copies/ml may still be contagious. Notably, the dynamics of transmission clusters made it possible to identify three diffusion zones, which predominantly differentiated between rural and urban areas, the latter being more prone to persistence and expansion, which may result in the emergence of new clusters nearby.
The use of geographic information is key for public health decision makers in mitigating the spread of the SARS-CoV-2 virus. This study suggests that early localization of clusters may help implement targeted protective measures limiting the spread of the virus.
Graphical abstract
Related collections
Most cited references53

- Record: found
- Abstract: found
- Article: found
Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR
- Record: found
- Abstract: found
- Article: not found