- Record: found
- Abstract: found
- Article: found
Adverse drug events and medication relation extraction in electronic health records with ensemble deep learning methods
Read this article at
Abstract
Objective
Identification of drugs, associated medication entities, and interactions among them are crucial to prevent unwanted effects of drug therapy, known as adverse drug events. This article describes our participation to the n2c2 shared-task in extracting relations between medication-related entities in electronic health records.
Materials and Methods
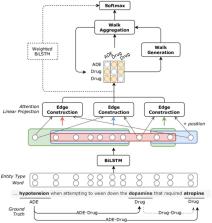
We proposed an ensemble approach for relation extraction and classification between drugs and medication-related entities. We incorporated state-of-the-art named-entity recognition (NER) models based on bidirectional long short-term memory (BiLSTM) networks and conditional random fields (CRF) for end-to-end extraction. We additionally developed separate models for intra- and inter-sentence relation extraction and combined them using an ensemble method. The intra-sentence models rely on bidirectional long short-term memory networks and attention mechanisms and are able to capture dependencies between multiple related pairs in the same sentence. For the inter-sentence relations, we adopted a neural architecture that utilizes the Transformer network to improve performance in longer sequences.
Results
Our team ranked third with a micro-averaged F1 score of 94.72% and 87.65% for relation and end-to-end relation extraction, respectively (Tracks 2 and 3). Our ensemble effectively takes advantages from our proposed models. Analysis of the reported results indicated that our proposed approach is more generalizable than the top-performing system, which employs additional training data- and corpus-driven processing techniques.
Conclusions
We proposed a relation extraction system to identify relations between drugs and medication-related entities. The proposed approach is independent of external syntactic tools. Analysis showed that by using latent Drug-Drug interactions we were able to significantly improve the performance of non–Drug-Drug pairs in EHRs.
Related collections
Most cited references11
- Record: found
- Abstract: found
- Article: not found
Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study.
- Record: found
- Abstract: found
- Article: not found
Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports.
- Record: found
- Abstract: found
- Article: not found