- Record: found
- Abstract: found
- Article: found
Reconstruction and analysis of circRNA-miRNA-mRNA network in the pathology of cervical cancer
Read this article at
Abstract
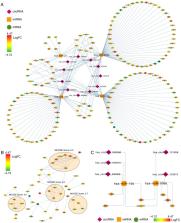
The present study was performed with the aim of understanding the mechanisms of pathogenesis and providing novel biomarkers for cervical cancer by constructing a regulatory circular (circ)RNA-micro (mi)RNA-mRNA network. Using an adjusted P-value of <0.05 and an absolute log value of fold-change >1, 16 and 156 miRNAs from GSE30656 and The Cancer Genome Atlas (TCGA), 5,321 mRNAs from GSE63514, 4,076 mRNAs from cervical squamous cell carcinoma and endocervical adenocarcinoma (from TCGA) and 75 circRNAs from GSE102686 were obtained. Using RNAhybrid, Venn and UpSetR plot, 12 circRNA-miRNA pairs and 266 miRNA-mRNA pairs were obtained. Once these pairs were combined, a circRNA-miRNA-mRNA network with 11 circRNA nodes, 4 miRNA nodes, 153 mRNA nodes and 203 edges was constructed. By constructing the protein-protein interaction network using Molecular Complex Detection scores >5 and >5 nodes, 7 hubgenes (RRM2, CEP55, CHEK1, KIF23, RACGAP1, ATAD2 and KIF11) were identified. By mapping the 7 hubgenes into the preliminary circRNA-miRNA-mRNA network, a circRNA-miRNA-hubgenes network consisting of 5 circRNAs (hsa_circRNA_000596, hsa_circRNA_104315, hsa_circRNA_400068, hsa_circRNA_101958 and hsa_circRNA_103519), 2 mRNAs (hsa-miR-15b and hsa-miR-106b) and 7 mRNAs (RRM2, CEP55, CHEK1, KIF23, RACGAP1, ATAD2 and KIF11) was constructed. There were 22 circRNA-miRNA-mRNA regulatory axes identified in the subnetwork. By analyzing the overall survival for the 7 hubgenes using the Gene Expression Profiling Interactive Analysis tool, higher expression of RRM2 was demonstrated to be associated with a significantly poorer overall survival. PharmGkb analysis identified single nucleotide polymorphisms (SNPs) of rs5030743 and rs1130609 of RRM2, which can be treated with cladribine and cytarabine. RRM2 was also indicated to be involved in the gemcitabine pathway. The 5 circRNAs (hsa_circRNA_000596, hsa_circRNA_104315, hsa_circRNA_400068, hsa_circRNA_101958 and hsa_circRNA_103519) may function as competing endogenous RNAs and serve critical roles in cervical cancer. In addition, cytarabine may produce similar effects to gemcitabine and may be an optional chemotherapeutic drug for treating cervical cancer by targeting rs5030743 and rs1130609 or other similar SNPs. However, the specific mechanism of action should be confirmed by further study.
Related collections
Most cited references44
- Record: found
- Abstract: found
- Article: not found
Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.
- Record: found
- Abstract: not found
- Article: not found
Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up.
- Record: found
- Abstract: found
- Article: not found