- Record: found
- Abstract: found
- Article: found
Atlas of breast cancer infiltrated B-lymphocytes revealed by paired single-cell RNA-sequencing and antigen receptor profiling
Read this article at
Abstract
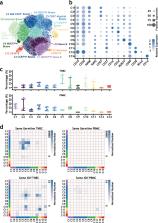
To gain mechanistic insights into the functions and developmental dynamics of tumor-infiltrated immune cells, especially B-lymphocytes, here we combine single-cell RNA-sequencing and antigen receptor lineage analysis to characterize a large number of triple-negative breast cancer infiltrated immune cells and report a comprehensive atlas of tumor-infiltrated B-lymphocytes. The single-cell transcriptional profiles reveal significant heterogeneity in tumor-infiltrated B-cell subgroups. The single-cell antigen receptor analyses demonstrate that compared with those in peripheral blood, tumor-infiltrated B-cells have more mature and memory B-cell characteristics, higher clonality, more class switching recombination and somatic hypermutations. Combined analyses suggest local differentiation of infiltrated memory B-cells within breast tumors. The B-cell signatures based on the single-cell RNA-sequencing results are significantly associated with improved survival in breast tumor patients. Functional analyses of tumor-infiltrated B-cell populations suggest that mechanistically, B-cell subgroups may contribute to immunosurveillance through various pathways. Further dissection of tumor-infiltrated B-cell populations will provide valuable clues for tumor immunotherapy.
Abstract
Immune cells infiltrating the tumour microenvironment play critical roles in disease pathogenesis and the immune response. Here the authors present the characterisation of infiltrating B cells in breast tumours by the formation of an atlas created from paired RNA sequence and antigen receptor profiling.
Related collections
Most cited references83
- Record: found
- Abstract: found
- Article: found
Hallmarks of Cancer: The Next Generation
- Record: found
- Abstract: found
- Article: not found
Integrating single-cell transcriptomic data across different conditions, technologies, and species

- Record: found
- Abstract: found
- Article: found