- Record: found
- Abstract: found
- Article: found
Multigene editing reveals that MtCEP1/ 2/ 12 redundantly control lateral root and nodule number in Medicago truncatula
Read this article at
Abstract
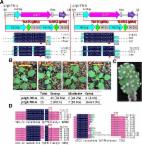
Multiple Mtcep mutants generated using a modified CRISPR/Cas9 system reveal the redundant roles of MtCEP1, 2, and 12 in controlling lateral root and nodule number in Medicago truncatula.
Abstract
The multimember CEP ( C-terminally Encoded Peptide) gene family is a complex group that is involved in various physiological activities in plants. Previous studies demonstrated that MtCEP1 and MtCEP7 control lateral root formation or nodulation, but these studies were based only on gain of function or artificial miRNA (amiRNA)/RNAi approaches, never knockout mutants. Moreover, an efficient multigene editing toolkit is not currently available for Medicago truncatula. Our quantitative reverse transcription–PCR data showed that MtCEP1, 2, 4, 5, 6, 7, 8, 9, 12, and 13 were up-regulated under nitrogen starvation conditions and that MtCEP1, 2, 7, 9, and 12 were induced by rhizobial inoculation. Treatment with synthetic MtCEP peptides of MtCEP1, 2, 4, 5, 6, 8, and 12 repressed lateral root emergence and promoted nodulation in the R108 wild type but not in the cra2 mutant. We optimized CRISPR/Cas9 [clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9] genome editing system for M. truncatula, and thus created single mutants of MtCEP1, 2, 4, 6, and 12 and the double mutants Mtcep1/2C and Mtcep5/8C; however, these mutants did not exhibit significant differences from R108. Furthermore, a triple mutant Mtcep1/2/12C and a quintuple mutant Mtcep1/2/5/8/12C were generated and exhibited more lateral roots and fewer nodules than R108. Overall, MtCEP1, 2, and 12 were confirmed to be redundantly important in the control of lateral root number and nodulation. Moreover, the CRISPR/Cas9-based multigene editing protocol provides an additional tool for research on the model legume M. truncatula, which is highly efficient at multigene mutant generation.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: found
A CRISPR/Cas9 toolkit for multiplex genome editing in plants
- Record: found
- Abstract: found
- Article: not found
The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti.
- Record: found
- Abstract: found
- Article: not found