- Record: found
- Abstract: found
- Article: found
Molecular mechanism of resveratrol promoting differentiation of preosteoblastic MC3T3-E1 cells based on network pharmacology and experimental validation
Read this article at
Abstract
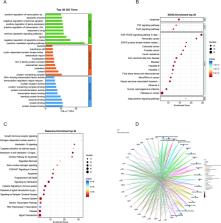
The purpose of this study was to investigate the mechanism by which resveratrol (Res) inhibits apoptosis and promotes proliferation and differentiation of pre-osteoblastic MC3T3-E1 cells, laying the groundwork for the treatment of osteoporosis (OP). The TCMSP database was used to find the gene targets for Res. The GeneCards database acquire the gene targets for OP. After discovering the potential target genes, GO, KEGG, and Reactome enrichment analysis were conducted. Verifying the major proteins involved in apoptosis can bind to Res using molecular docking. CCK8 measured the proliferative activity of mouse pre-osteoblasts in every group following Res intervention. Alkaline phosphatase staining (ALP) and alizarin red staining to measure the ability of osteogenic differentiation. RT-qPCR to determine the expression levels of Runx2 and OPG genes for osteogenic differentiation ability of cells. Western blot to measure the degree of apoptosis-related protein activity in each group following Res intervention. The biological processes investigated for GO of Res therapeutic OP involved in cytokine-mediated signaling pathway, negative regulation of apoptotic process, Aging, extrinsic apoptotic signaling pathway in absence of ligand, according to potential therapeutic target enrichment study. Apoptosis, FoxO signaling pathway, and TNF signaling pathway are the primary KEGG signaling pathways. Recactome pathways are primarily engaged in Programmed Cell Death, Apoptosis, Intrinsic Apoptotic Pathway, and Caspase activation via extrinsic apoptotic signaling pathways. This research established a new approach for Res treatment of OP by demonstrating how Res controls the apoptosis-related proteins TNF, IL6, and CASP3 to suppress osteoblast death and increase osteoclastogenesis.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Cytoscape: a software environment for integrated models of biomolecular interaction networks.
- Record: found
- Abstract: found
- Article: not found
KEGG: kyoto encyclopedia of genes and genomes.
- Record: found
- Abstract: found
- Article: not found