- Record: found
- Abstract: found
- Article: found
Specific versus Non-Specific Immune Responses in an Invertebrate Species Evidenced by a Comparative de novo Sequencing Study
Read this article at
Abstract
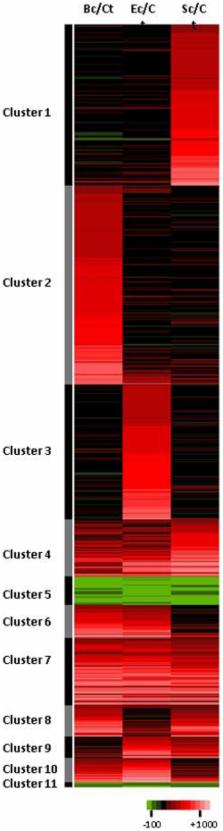
Our present understanding of the functioning and evolutionary history of invertebrate innate immunity derives mostly from studies on a few model species belonging to ecdysozoa. In particular, the characterization of signaling pathways dedicated to specific responses towards fungi and Gram-positive or Gram-negative bacteria in Drosophila melanogaster challenged our original view of a non-specific immunity in invertebrates. However, much remains to be elucidated from lophotrochozoan species. To investigate the global specificity of the immune response in the fresh-water snail Biomphalaria glabrata, we used massive Illumina sequencing of 5′-end cDNAs to compare expression profiles after challenge by Gram-positive or Gram-negative bacteria or after a yeast challenge. 5′-end cDNA sequencing of the libraries yielded over 12 millions high quality reads. To link these short reads to expressed genes, we prepared a reference transcriptomic database through automatic assembly and annotation of the 758,510 redundant sequences (ESTs, mRNAs) of B. glabrata available in public databases. Computational analysis of Illumina reads followed by multivariate analyses allowed identification of 1685 candidate transcripts differentially expressed after an immune challenge, with a two fold ratio between transcripts showing a challenge-specific expression versus a lower or non-specific differential expression. Differential expression has been validated using quantitative PCR for a subset of randomly selected candidates. Predicted functions of annotated candidates (approx. 700 unisequences) belonged to a large extend to similar functional categories or protein types. This work significantly expands upon previous gene discovery and expression studies on B. glabrata and suggests that responses to various pathogens may involve similar immune processes or signaling pathways but different genes belonging to multigenic families. These results raise the question of the importance of gene duplication and acquisition of paralog functional diversity in the evolution of specific invertebrate immune responses.
Related collections
Most cited references73
- Record: found
- Abstract: found
- Article: not found
Melanogenesis and associated cytotoxic reactions: applications to insect innate immunity.
- Record: found
- Abstract: found
- Article: not found
Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae.
- Record: found
- Abstract: found
- Article: not found