- Record: found
- Abstract: found
- Article: found
Memory T Cells in Latent Mycobacterium tuberculosis Infection Are Directed against Three Antigenic Islands and Largely Contained in a CXCR3 +CCR6 + Th1 Subset
Read this article at
Abstract
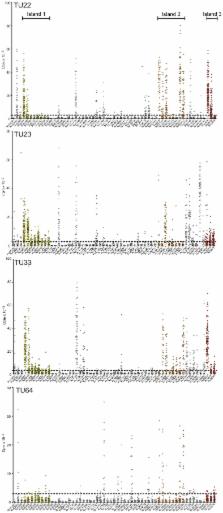
An understanding of the immunological footprint of Mycobacterium tuberculosis (MTB) CD4 T cell recognition is still incomplete. Here we report that human Th1 cells specific for MTB are largely contained in a CXCR3 +CCR6 + memory subset and highly focused on three broadly immunodominant antigenic islands, all related to bacterial secretion systems. Our results refute the notion that secreted antigens act as a decoy, since both secreted proteins and proteins comprising the secretion system itself are targeted by a fully functional T cell response. In addition, several novel T cell antigens were identified which can be of potential diagnostic use, or as vaccine antigens. These results underline the power of a truly unbiased, genome-wide, analysis of CD4 MTB recognition based on the combined use of epitope predictions, high throughput ELISPOT, and T cell libraries using PBMCs from individuals latently infected with MTB.
Author Summary
Mycobacterium tuberculosis is one of the most life-threatening pathogens of all time, having infected one-third of the present human population. There is an urgent need for both novel vaccines and diagnostic strategies. Here, we were able to identify the targets most dominantly recognized by latently infected individual that successfully contain infection. These targets are contained in three broadly genomic antigenic islands, all related to bacterial secretion systems and composed by several distinct ORFs. Thus, our results suggest that vaccination with one or few defined antigens will fail to replicate the response associated with natural immunity. Our analysis also pinpoints that the Th1 cells dominating the response are associated with novel and well-defined phenotypic markers, suggesting that the response is molded by unique MTB associated factors. This study demonstrates further that the approach combining peptide binding predictions with modern high throughput techniques is generally applicable to the study of immunity to other complex pathogens. Together, our data provide a new angle in the worldwide fight against M. tuberculosis and could be used for diagnostic or vaccine developments.
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
Genetic requirements for mycobacterial survival during infection.
- Record: found
- Abstract: found
- Article: not found
Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells.
- Record: found
- Abstract: found
- Article: not found