- Record: found
- Abstract: found
- Article: found
Ex vivo prime editing of patient haematopoietic stem cells rescues sickle-cell disease phenotypes after engraftment in mice
Read this article at
Abstract
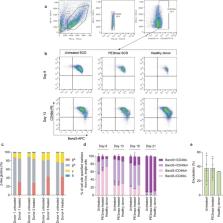
Sickle-cell disease (SCD) is caused by an A·T-to-T·A transversion mutation in the β-globin gene ( HBB). Here we show that prime editing can correct the SCD allele ( HBB S) to wild type ( HBB A) at frequencies of 15%–41% in haematopoietic stem and progenitor cells (HSPCs) from patients with SCD. Seventeen weeks after transplantation into immunodeficient mice, prime-edited SCD HSPCs maintained HBB A levels and displayed engraftment frequencies, haematopoietic differentiation and lineage maturation similar to those of unedited HSPCs from healthy donors. An average of 42% of human erythroblasts and reticulocytes isolated 17 weeks after transplantation of prime-edited HSPCs from four SCD patient donors expressed HBB A, exceeding the levels predicted for therapeutic benefit. HSPC-derived erythrocytes carried less sickle haemoglobin, contained HBB A -derived adult haemoglobin at 28%–43% of normal levels and resisted hypoxia-induced sickling. Minimal off-target editing was detected at over 100 sites nominated experimentally via unbiased genome-wide analysis. Our findings support the feasibility of a one-time prime editing SCD treatment that corrects HBB S to HBB A, does not require any viral or non-viral DNA template and minimizes undesired consequences of DNA double-strand breaks.
Abstract
Prime editing can efficiently correct the sickle-cell allele to produce wild-type haemoglobin in patient haematopoietic stem cells that engraft efficiently in mice, yielding erythrocytes resistant to hypoxia-induced sickling.
Related collections
Most cited references64
- Record: found
- Abstract: found
- Article: not found
Search-and-replace genome editing without double-strand breaks or donor DNA
- Record: found
- Abstract: found
- Article: not found
Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements
- Record: found
- Abstract: found
- Article: not found