- Record: found
- Abstract: found
- Article: found
Maintenance of human haematopoietic stem and progenitor cells in vitro using a chemical cocktail
Read this article at
Abstract
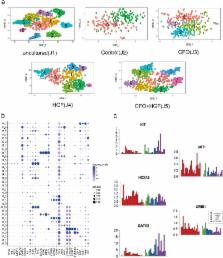
Identification of effective culture conditions to maintain and possibly expand human HSPCs in vitro is an important goal. Recent advances highlight the efficacy of chemicals in maintaining and converting cell fates. We screened 186 chemicals and found that a combination of CHIR-99021, Forskolin and OAC1 (CFO) maintained human CD34-positive cells in vitro. Efficiency of the culture system was characterized using flow cytometry for CD34-positive cells, a colony-forming assay and xeno-transplants. We found that human CD34-positive cells treated with this combination had enhanced expression of human HSPC markers and increased haematopoietic re-populating ability in immune-deficient mice. Single-cell RNA-seq analyses showed that the in vitro cultured human CD34-positive cells were heterogeneous. We found that CFO supports maintenance of human CD34-positive cells by activating HOXA9, GATA2 and AKT-cAMP signaling pathway. These data have implications in therapies requiring maintenance and/or expansion of human HSPCs.
Related collections
Most cited references18
- Record: found
- Abstract: found
- Article: not found
Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells.
- Record: found
- Abstract: found
- Article: not found
Distinct routes of lineage development reshape the human blood hierarchy across ontogeny.
- Record: found
- Abstract: found
- Article: not found