- Record: found
- Abstract: found
- Article: found
Comparative lipidome and transcriptome provide novel insights into zero-valent iron nanoparticle-treated Fremyella diplosiphon
Read this article at
Abstract
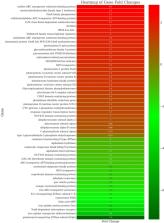
Understanding the intricate interplay between nanoparticle-mediated cyanobacterial interactions is pivotal in elucidating their impact on the transcriptome and lipidome. In the present study, total fatty acid methyl esters (FAMEs) in the wild-type (B481-WT) and transformant (B481-SD) Fremyella diplosiphon strains treated with nanoscale zero-valent iron nanoparticles (nZVIs) were characterized, and transcriptome changes analyzed. Comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry revealed a 20–25% higher percentage of FAMEs in nZVI-treated F. diplosiphon strain B481-SD compared to B481-WT. Accumulation of alkanes was significantly higher (> 1.4 times) in both strains treated with 25.6 mg L −1 nZVIs compared to the untreated control. In addition, we observed significantly higher levels of monounsaturated FAMEs (11%) in B481-WT in 3.2 (11.34%) and 25.6 mg L −1 (11.22%) nZVI-treated cells when compared to the untreated control (7%). Analysis of the F. diplosiphon transcriptome treated with 3.2 mg L −1 revealed a total of 1811 and 1651 genes that were differentially expressed in B481-SD and B481-WT respectively. While the expression of iron uptake and ion channel genes was downregulated, genes coding for photosynthesis, pigment, and antioxidant enzymes were significantly ( p < 0.05) upregulated in B481-SD treated with 3.2 mg L −1 nZVIs compared to the untreated control. This study on essential FAMEs and regulation of genes in nZVI-treated F. diplosiphon strains provides a molecular framework for optimization of metabolic pathways in this model species.
Related collections
Most cited references55
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.
- Record: found
- Abstract: found
- Article: not found