- Record: found
- Abstract: found
- Article: not found
circRNA expression patterns and circRNA-miRNA-mRNA networks during CV-A16 infection of SH-SY5Y cells
Read this article at
Abstract
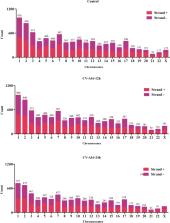
Coxsackievirus A16 (CV-A16) has caused worldwide epidemics of hand, foot, and mouth disease (HFMD) in infants and preschool children. Circular RNAs (circRNAs), a class of noncoding RNA molecules, participate in the progression of viral infectious diseases. Although the function of circRNAs has been a heavily researched topic, their role in CV-A16 infection is still unclear. In this study, the viral effects of CV-A16 on the cellular circRNA transcriptome were investigated using next-generation sequencing technology. The results showed that a total of 8726, 8611, and 6826 circRNAs were identified at 0, 12, and 24 h postinfection, respectively. Moreover, it was found that 1769 and 1192 circRNAs were differentially expressed in at 12 and 24 h postinfection, respectively. The common differentially expressed circRNAs were used for functional annotation analysis, and it was found that the parent genes of differentially expressed circRNAs might be associated with the viral infection process, especially the “Immune system process” in GO analysis and the “Inflammation mediated by chemokine and cytokine signaling pathway” in KEGG analysis. Subsequently, circRNA-miRNA-mRNA regulatory networks were constructed, and the hsa_circ_0004447/hsa-miR-942-5p/MMP2, hsa_circ_0078617/hsa-miR-6780b-5p/MMP2 and hsa_circ_0078617/hsa-miR-5196-5p/MMP2 regulatory axes were identified by enrichment analysis as important networks during the progression of CV-A16 infection. Finally, six dysregulated circRNAs were selected for validation and were verified to be consistent with the sequencing results. Considering all of these results, to the best of our knowledge, this study is the first to present a comprehensive overview of circRNAs induced by CV-A16 infection, and this research demonstrated that a network of enriched circRNAs and circRNA-associated competitive endogenous RNAs (ceRNAs) is involved in the regulation of CV-A16 infection, thereby helping to elucidate the mechanisms underlying CV-A16-host interactions.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
The biogenesis, biology and characterization of circular RNAs
- Record: found
- Abstract: found
- Article: not found
The multilayered complexity of ceRNA crosstalk and competition.
- Record: found
- Abstract: found
- Article: not found
