- Record: found
- Abstract: found
- Article: found
Natural mating ability is associated with gut microbiota composition and function in captive male giant pandas
Read this article at
Abstract
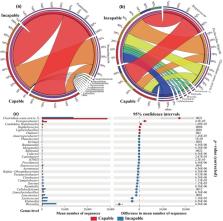
The issue of poor sexual performance of some male giant pandas seriously impairs the growth and the genetic diversity of the captive population, yet there is still no clear understanding of the cause of the loss of this ability and its underlying mechanism. In this study, we analyzed the gut microbiota and its function in 72 fecal samples obtained from 20 captive male giant pandas, with an equal allocation between individuals capable and incapable of natural mating. Additionally, we investigated fecal hormone levels and behavioral differences between the two groups. A correlation analysis was then conducted among these factors to explore the influencing factors of their natural mating ability. The results showed significant differences in the composition of gut microbiota between the two groups of male pandas. The capable group had significantly higher abundance of Clostridium sensu stricto 1 ( p adjusted = .0021, GLMM), which was positively correlated with fatty acid degradation and two‐component system functions (Spearman, p adjusted < .05). Additionally, the capable group showed higher gene abundance in gut microbiota function including purine and pyrimidine metabolism and galactose metabolism, as well as pathways related to biological processes such as ribosome and homologous recombination (DEseq2, p adjusted < .05). We found no significant differences in fecal cortisol and testosterone levels between the two groups, and no difference was found in their behavior either. Our study provides a theoretical and practical basis for further studying the behavioral degradation mechanisms of giant pandas and other endangered mammal species.
Abstract
This study investigated the gut microbiota of captive male giant pandas and found significant differences between the microbiota of individuals’ capable and incapable of natural mating. The findings provide insight into potential mechanisms affecting the reproductive success of endangered mammal species.
Related collections
Most cited references96
- Record: found
- Abstract: found
- Article: not found
DADA2: High resolution sample inference from Illumina amplicon data

- Record: found
- Abstract: found
- Article: found
phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data
- Record: found
- Abstract: found
- Article: found
