- Record: found
- Abstract: found
- Article: found
The profiling of microbiota in vaginal swab samples using 16S rRNA gene sequencing and IS-pro analysis
Read this article at
Abstract
Background
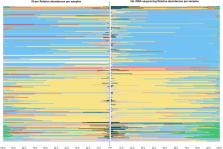
16S rRNA gene sequencing is currently the most common way of determining the composition of microbiota. This technique has enabled many new discoveries to be made regarding the relevance of microbiota to the health and disease of the host. However, compared to other diagnostic techniques, 16S rRNA gene sequencing is fairly costly and labor intensive, leaving room for other techniques to improve on these aspects.
Results
The current study aimed to compare the output of 16S rRNA gene sequencing to the output of the quick IS-pro analysis, using vaginal swab samples from 297 women of reproductive age. 16S rRNA gene sequencing and IS-pro analyses yielded very similar vaginal microbiome profiles, with a median Pearson’s R 2 of 0.97, indicating a high level of similarity between both techniques.
Related collections
Most cited references16
- Record: found
- Abstract: found
- Article: not found
Search and clustering orders of magnitude faster than BLAST.
- Record: found
- Abstract: found
- Article: not found
Vaginal microbiome of reproductive-age women.

- Record: found
- Abstract: found
- Article: found