- Record: found
- Abstract: found
- Article: found
PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data
Read this article at
Abstract
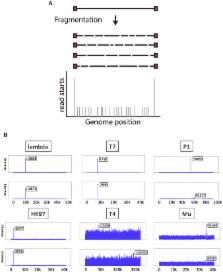
The worrying rise of antibiotic resistance in pathogenic bacteria is leading to a renewed interest in bacteriophages as a treatment option. Novel sequencing technologies enable description of an increasing number of phage genomes, a critical piece of information to understand their life cycle, phage-host interactions, and evolution. In this work, we demonstrate how it is possible to recover more information from sequencing data than just the phage genome. We developed a theoretical and statistical framework to determine DNA termini and phage packaging mechanisms using NGS data. Our method relies on the detection of biases in the number of reads, which are observable at natural DNA termini compared with the rest of the phage genome. We implemented our method with the creation of the software PhageTerm and validated it using a set of phages with well-established packaging mechanisms representative of the termini diversity, i.e. 5′ cos (Lambda), 3′ cos (HK97), pac (P1), headful without a pac site (T4), DTR (T7) and host fragment (Mu). In addition, we determined the termini of nine Clostridium difficile phages and six phages whose sequences were retrieved from the Sequence Read Archive. PhageTerm is freely available (https://sourceforge.net/projects/phageterm), as a Galaxy ToolShed and on a Galaxy-based server (https://galaxy.pasteur.fr).
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.

- Record: found
- Abstract: found
- Article: found
Phamerator: a bioinformatic tool for comparative bacteriophage genomics
- Record: found
- Abstract: not found
- Article: not found