- Record: found
- Abstract: found
- Article: found
The prognostic value of B7-H6 in esophageal squamous cell carcinoma
Read this article at
Abstract
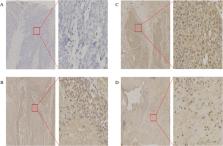
B7-H6, a member of the B7 family molecules, participates in the clearance of tumor cells by binding to NKp30 on NK cells. B7-H6 expression level in esophageal squamous cell carcinoma (ESCC) and the clinical value remain unknown. The goal of this study was to determine the expression of B7-H6 in ESCC and further explore its clinical significance. We retrospectively collected the clinical data of 145 patients diagnosed with ESCC between January 2007 and December 2008. The expression of B7-H6 of the pathological tissue samples was detected by immunohistochemistry. The chi-square (χ 2) test was used to analyse the relationships of B7-H6 and clinicopathological characteristics. Survival and hazard functions were estimated using the Kaplan-Meier method, and survival between groups was compared using the two-sided log-rank test. The Cox proportional hazards regression model was used to adjust for the risk factors related to overall survival (OS). 133/145 (91.72%) of the ESCC tissue samples exhibited B7-H6 expression. The expression level of B7-H6 was correlated with T stage ( P = 0.036) and lymphatic metastasis status ( P = 0.044). High B7-H6 expression ( P = 0.003) was associated with a significantly worse OS than low B7-H6 expression. Multivariate Cox proportional hazards regression analysis demonstrated that tumour size ( P = 0.021), B7-H6 expression ( P = 0.025) and lymphatic metastasis status ( P = 0.049) were independent prognostic factors of OS for ESCC. Collectively, our findings suggest that B7-H6 is widely expressed in ESCC samples. And B7-H6 may represent a predictor of poor prognosis for ESCC.
Related collections
Most cited references29

- Record: found
- Abstract: found
- Article: found
The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans
- Record: found
- Abstract: found
- Article: not found