- Record: found
- Abstract: found
- Article: found
Discovery and optimized extraction of the anti-osteoclastic agent epicatechin-7-O-β-D-apiofuranoside from Ulmus macrocarpa Hance bark

Read this article at
Abstract
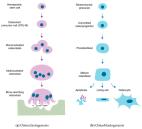
Ulmus macrocarpa Hance bark ( UmHb) has been used as a traditional herbal medicine in East Asia for bone concern diseases for a long time. To find a suitable solvent, we, in this study, compared the efficacy of UmHb water extract and ethanol extract which can inhibit osteoclast differentiation. Compared with two ethanol extracts (70% and 100% respectively), hydrothermal extracts of UmHb more effectively inhibited receptor activators of nuclear factor κB ligand-induced osteoclast differentiation in murine bone marrow-derived macrophages. We identified for the first time that (2R,3R)-epicatechin-7-O-β-D-apiofuranoside (E7A) is a specific active compound in UmHb hydrothermal extracts through using LC/MS, HPLC, and NMR techniques. In addition, we confirmed through TRAP assay, pit assay, and PCR assay that E7A is a key compound in inhibiting osteoclast differentiation. The optimized condition to obtain E7A-rich UmHb extract was 100 mL/g, 90 °C, pH 5, and 97 min. At this condition, the content of E7A was 26.05 ± 0.96 mg/g extract. Based on TRAP assay, pit assay, PCR, and western blot, the optimized extract of E7A-rich UmHb demonstrated a greater inhibition of osteoclast differentiation compared to unoptimized. These results suggest that E7A would be a good candidate for the prevention and treatment of osteoporosis-related diseases.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Control of osteoblast function and regulation of bone mass.
- Record: found
- Abstract: found
- Article: not found
Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts.
- Record: found
- Abstract: found
- Article: found