- Record: found
- Abstract: found
- Article: found
Circular RNA hsa_circ_0043688 serves as a competing endogenous RNA for microRNA‐145‐5p to promote the progression of Keloids via Fibroblast growth factor‐2
Read this article at
Abstract
Background
Keloids are benign fibroproliferative skin tumors. Circular RNA (circRNA) hsa_circ_0043688 has been exhibited to the freakishly expressed in keloid tissues. Here, we aimed to investigate the regulatory network of hsa_circ_0043688 in the pathological process of keloid.
Methods
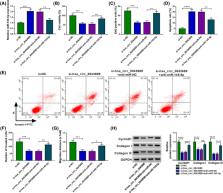
Hsa_circ_0043688, microRNA‐145‐5p (miR‐145‐5p), and Fibroblast growth factor‐2 (FGF2) level were detected using RT‐qPCR. Cell viability, proliferation, apoptosis, invasion, and migration were investigated using Cell Counting Kit‐8 (CCK‐8), 5‐ethynyl‐2′‐deoxyuridine (EdU), flow cytometry, transwell, and wound healing assays, respectively. Western blot analysis of protein levels of FGF2, CyclinD1, Collagen I, and Collagen III. After the prediction of Circinteractome and Starbase, their interaction was verified based on a dual‐luciferase reporter and RIP assays.
Results
Increased hsa_circ_0043688 and FGF2, and decreased miR‐145‐5p in keloids samples and fibroblasts were found. Also, hsa_circ_0043688 absence hindered proliferation, invasion, migration, and boost apoptosis of keloid fibroblasts. In mechanism, hsa_circ_0043688 modulated FGF2 content via sponging miR‐145‐5p.
Abstract
Related collections
Most cited references36
- Record: found
- Abstract: found
- Article: not found
Natural RNA circles function as efficient microRNA sponges.
- Record: found
- Abstract: found
- Article: not found