- Record: found
- Abstract: found
- Article: found
Effect of sublethal dose of chloramphenicol on biofilm formation and virulence in Vibrio parahaemolyticus
Read this article at
Abstract
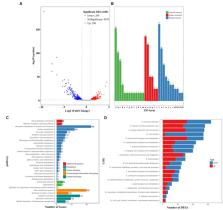
Vibrio parahaemolyticus isolates are generally very sensitive to chloramphenicol. However, it is usually necessary to transfer a plasmid carrying a chloramphenicol resistance gene into V. parahaemolyticus to investigate the function of a specific gene, and the effects of chloramphenicol on bacterial physiology have not been investigated. In this work, the effects of sublethal dose of chloramphenicol on V. parahaemolyticus were investigated by combined utilization of various phenotypic assays and RNA sequencing (RNA-seq). The results showed that the growth rate, biofilm formation capcity, c-di-GMP synthesis, motility, cytoxicity and adherence activity of V. parahaemolyticus were remarkably downregulated by the sublethal dose of chloramphenicol. The RNA-seq data revealed that the expression levels of 650 genes were significantly differentially expressed in the response to chloramphenicol stress, including antibiotic resistance genes, major virulence genes, biofilm-associated genes and putative regulatory genes. Majority of genes involved in the synthesis of polar flagellum, exopolysaccharide (EPS), mannose-sensitive haemagglutinin type IV pilus (MSHA), type III secretion systems (T3SS1 and T3SS2) and type VI secretion system 2 (T6SS2) were downregulated by the sublethal dose of chloramphenicol. Five putative c-di-GMP metabolism genes were significantly differentially expressed, which may be the reason for the decrease in intracellular c-di-GMP levels in the response of chloramphenicol stress. In addition, 23 genes encoding putative regulators were also significantly differentially expressed, suggesting that these regulators may be involved in the resistance of V. parahaemolyticus to chloramphenicol stress. This work helps us to understand how chloramphenicol effect on the physiology of V. parahaemolyticus.
Related collections
Most cited references98
- Record: found
- Abstract: found
- Article: not found
Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter.
- Record: found
- Abstract: found
- Article: not found
Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae.
- Record: found
- Abstract: found
- Article: not found