- Record: found
- Abstract: found
- Article: found
HLA Class I and Class II Conserved Extended Haplotypes and Their Fragments or Blocks in Mexicans: Implications for the Study of Genetic Diversity in Admixed Populations
Read this article at
Abstract
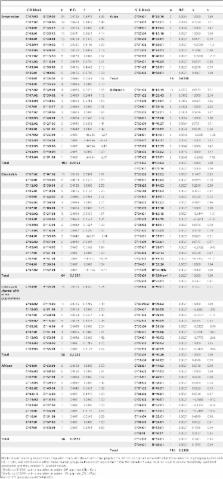
Major histocompatibility complex (MHC) genes are highly polymorphic and informative in disease association, transplantation, and population genetics studies with particular importance in the understanding of human population diversity and evolution. The aim of this study was to describe the HLA diversity in Mexican admixed individuals. We studied the polymorphism of MHC class I ( HLA-A, -B, -C), and class II ( HLA-DRB1, -DQB1) genes using high-resolution sequence based typing (SBT) method and we structured the blocks and conserved extended haplotypes (CEHs) in 234 non-related admixed Mexican individuals (468 haplotypes) by a maximum likelihood method. We found that HLA blocks and CEHs are primarily from Amerindian and Caucasian origin, with smaller participation of African and recent Asian ancestry, demonstrating a great diversity of HLA blocks and CEHs in Mexicans from the central area of Mexico. We also analyzed the degree of admixture in this group using short tandem repeats (STRs) and HLA-B that correlated with the frequency of most probable ancestral HLA-C/− B and -DRB1/− DQB1 blocks and CEHs. Our results contribute to the analysis of the diversity and ancestral contribution of HLA class I and HLA class II alleles and haplotypes of Mexican admixed individuals from Mexico City. This work will help as a reference to improve future studies in Mexicans regarding allotransplantation, immune responses and disease associations.
Related collections
Most cited references69
- Record: found
- Abstract: found
- Article: found
Arlequin (version 3.0): An integrated software package for population genetics data analysis
- Record: found
- Abstract: found
- Article: not found
Gene map of the extended human MHC.
- Record: found
- Abstract: found
- Article: not found