- Record: found
- Abstract: found
- Article: found
Evidence that a West-East admixed population lived in the Tarim Basin as early as the early Bronze Age
Read this article at
Abstract
Background
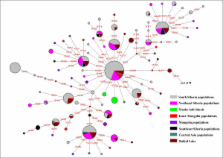
The Tarim Basin, located on the ancient Silk Road, played a very important role in the history of human migration and cultural communications between the West and the East. However, both the exact period at which the relevant events occurred and the origins of the people in the area remain very obscure. In this paper, we present data from the analyses of both Y chromosomal and mitochondrial DNA (mtDNA) derived from human remains excavated from the Xiaohe cemetery, the oldest archeological site with human remains discovered in the Tarim Basin thus far.
Results
Mitochondrial DNA analysis showed that the Xiaohe people carried both the East Eurasian haplogroup (C) and the West Eurasian haplogroups (H and K), whereas Y chromosomal DNA analysis revealed only the West Eurasian haplogroup R1a1a in the male individuals.
Conclusion
Our results demonstrated that the Xiaohe people were an admixture from populations originating from both the West and the East, implying that the Tarim Basin had been occupied by an admixed population since the early Bronze Age. To our knowledge, this is the earliest genetic evidence of an admixed population settled in the Tarim Basin.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Technical note: improved DNA extraction from ancient bones using silica-based spin columns.
- Record: found
- Abstract: found
- Article: not found
The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective.
- Record: found
- Abstract: found
- Article: not found