- Record: found
- Abstract: found
- Article: found
Intracellular calcium links milk stasis to lysosome-dependent cell death during early mammary gland involution
Read this article at
Abstract
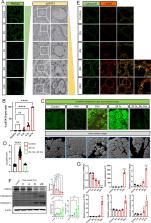
Involution of the mammary gland after lactation is a dramatic example of coordinated cell death. Weaning causes distension of the alveolar structures due to the accumulation of milk, which, in turn, activates STAT3 and initiates a caspase-independent but lysosome-dependent cell death (LDCD) pathway. Although the importance of STAT3 and LDCD in early mammary involution is well established, it has not been entirely clear how milk stasis activates STAT3. In this report, we demonstrate that protein levels of the PMCA2 calcium pump are significantly downregulated within 2–4 h of experimental milk stasis. Reductions in PMCA2 expression correlate with an increase in cytoplasmic calcium in vivo as measured by multiphoton intravital imaging of GCaMP6f fluorescence. These events occur concomitant with the appearance of nuclear pSTAT3 expression but prior to significant activation of LDCD or its previously implicated mediators such as LIF, IL6, and TGFβ3, all of which appear to be upregulated by increased intracellular calcium. We further demonstrate that increased intracellular calcium activates STAT3 by inducing degradation of its negative regulator, SOCS3. We also observed that milk stasis, loss of PMCA2 expression and increased intracellular calcium levels activate TFEB, an important regulator of lysosome biogenesis through a process involving inhibition of CDK4/6 and cell cycle progression. In summary, these data suggest that intracellular calcium serves as an important proximal biochemical signal linking milk stasis to STAT3 activation, increased lysosomal biogenesis, and lysosome-mediated cell death.
Related collections
Most cited references63

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
HISAT: a fast spliced aligner with low memory requirements.

- Record: found
- Abstract: found
- Article: found