- Record: found
- Abstract: found
- Article: found
Single Cell–ICP–ToF-MS for the Multiplexed Determination of Proteins: Evaluation of the Cellular Stress Response

Read this article at
Abstract
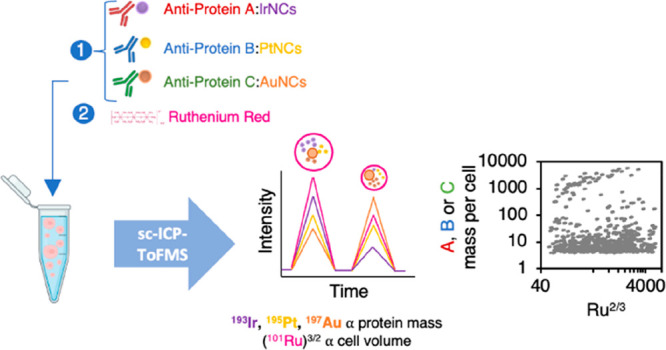
An automated and straightforward detection and data treatment strategy for the determination of the protein relative concentration in individual human cells by single cell–inductively coupled plasma–time-of-flight mass spectrometry (sc-ICP-ToF-MS) is proposed. Metal nanocluster (NC)-labeled specific antibodies for the target proteins were employed, and ruthenium red (RR) staining, which binds to the cells surface, was used to determine the number of cell events as well as to evaluate the relative volume of the cells. As a proof of concept, the expression of hepcidin, metallothionein-2, and ferroportin employing specific antibodies labeled with IrNCs, PtNCs, and AuNCs, respectively, was investigated by sc-ICP-ToF-MS in human ARPE-19 cells. Taking into account that ARPE-19 cells are spherical in suspension and RR binds to the surface of the cells, the Ru intensity was related to the cell volume (i.e., the cell volume is directly proportional to (Ru intensity) 3/2), making it possible to determine not only the mass of the target proteins in each individual cell but also the relative concentration. The proposed approach is of particular interest in comparing cell cultures subjected to different supplementations. ARPE-19 cell cultures under two stress conditions were compared: a hyperglycemic model and an oxidative stress model. The comparison of the control with treated cells shows not only the mass of analyzed species but also the relative changes in the cell volume and concentration of target proteins, clearly allowing the identification of subpopulations under the respective treatment.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Single-cell sequencing-based technologies will revolutionize whole-organism science.
- Record: found
- Abstract: found
- Article: not found
Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells.
- Record: found
- Abstract: found
- Article: not found
Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms.
Author and article information
Comments
Comment on this article
 Smart Citations
Smart CitationsSee how this article has been cited at scite.ai
scite shows how a scientific paper has been cited by providing the context of the citation, a classification describing whether it supports, mentions, or contrasts the cited claim, and a label indicating in which section the citation was made.