- Record: found
- Abstract: found
- Article: found
Insight into the Mechanisms of Adenovirus Capsid Disassembly from Studies of Defensin Neutralization
Read this article at
Abstract
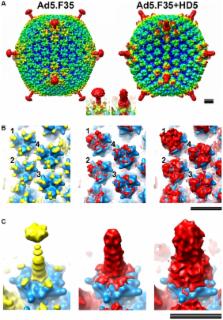
Defensins are effectors of the innate immune response with potent antibacterial activity. Their role in antiviral immunity, particularly for non-enveloped viruses, is poorly understood. We recently found that human alpha-defensins inhibit human adenovirus (HAdV) by preventing virus uncoating and release of the endosomalytic protein VI during cell entry. Consequently, AdV remains trapped in the endosomal/lysosomal pathway rather than trafficking to the nucleus. To gain insight into the mechanism of defensin-mediated neutralization, we analyzed the specificity of the AdV-defensin interaction. Sensitivity to alpha-defensin neutralization is a common feature of HAdV species A, B1, B2, C, and E, whereas species D and F are resistant. Thousands of defensin molecules bind with low micromolar affinity to a sensitive serotype, but only a low level of binding is observed to resistant serotypes. Neutralization is dependent upon a correctly folded defensin molecule, suggesting that specific molecular interactions occur with the virion. CryoEM structural studies and protein sequence analysis led to a hypothesis that neutralization determinants are located in a region spanning the fiber and penton base proteins. This model was supported by infectivity studies using virus chimeras comprised of capsid proteins from sensitive and resistant serotypes. These findings suggest a mechanism in which defensin binding to critical sites on the AdV capsid prevents vertex removal and thereby blocks subsequent steps in uncoating that are required for release of protein VI and endosomalysis during infection. In addition to informing the mechanism of defensin-mediated neutralization of a non-enveloped virus, these studies provide insight into the mechanism of AdV uncoating and suggest new strategies to disrupt this process and inhibit infection.
Author Summary
Defensins are effectors of the innate immune response with antibacterial and antiviral activity. A major bactericidal mechanism of defensins is membrane disruption; however, their mechanism against non-enveloped viruses, such as human adenovirus, is poorly understood. This work shows that sensitivity of human adenovirus to defensins is species specific and that neutralization is dependent upon defensin tertiary structure. A cryoEM structural study of an adenovirus vector in complex with a neutralizing defensin, HD5, led to a neutralization model in which defensin binds to the interface of two capsid proteins, preventing dissociation of the fiber protein. We propose that binding at this site blocks downstream uncoating events required for infection. Infectivity studies using virus chimeras comprised of capsid proteins from sensitive and resistant human adenovirus serotypes support this model. This functional and structural study provides insight into the mechanism of human adenovirus neutralization by defensins and suggests new strategies to inhibit infection.
Related collections
Most cited references54
- Record: found
- Abstract: found
- Article: not found
Defensins: antimicrobial peptides of innate immunity.
- Record: found
- Abstract: found
- Article: not found
Defensins. Natural peptide antibiotics of human neutrophils.
- Record: found
- Abstract: found
- Article: not found