- Record: found
- Abstract: found
- Article: found
Machine Learning With K-Means Dimensional Reduction for Predicting Survival Outcomes in Patients With Breast Cancer
Read this article at
Abstract
Objective:
Despite existing prognostic markers, breast cancer prognosis remains a difficult subject due to the complex relationships between many contributing factors and survival. This study seeks to integrate multiple clinicopathological and genomic factors with dimensional reduction across machine learning algorithms to compare survival predictions.
Methods:
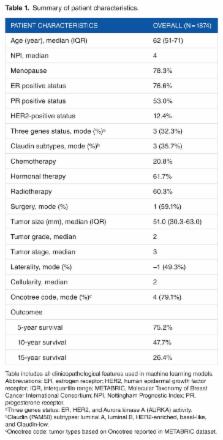
This is a secondary analysis of the data from a prospective cohort study of female patients with breast cancer enrolled in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC). We constructed a series of predictive models: ensemble models (Gradient Boosting and Random Forest), support vector machine (SVM), and artificial neural networks (ANN) for 5-year survival based on clinicopathological and gene expression data after K-means clustering with K-nearest-neighbor (KNN) classification. Model performance was evaluated by receiver operating characteristic (ROC) curve, accuracy, and calibration slope (CS). Model stability was assessed over 10 random runs in terms of ROC, accuracy, CS, and variable importance.
Results:
The analytic cohort is composed of 1874 patients with breast cancer. Overall, the median age was 62 years; the 5-year survival rate was 75%. ROC and accuracy were not significantly different between models (ROC and accuracy around 0.67 and 0.72 across models, respectively). However, ensemble methods resulted in better fit (CS) with stable measures of variable importance across 10 random training/validation splits. K-means clustering of gene expression profiles on training data points along with KNN classification of validation data points was a robust method of dimensional reduction. Furthermore, the gene expression cluster with the highest mortality risk was an influential factor in model prediction.
Conclusions:
Using machine learning methods to construct predictive models for 5-year survival in patients with breast cancer, we demonstrated discrimination ability across models with new insight into the stability and utility of dimensional reduction on genomic features in breast cancer survival prediction.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal.
- Record: found
- Abstract: found
- Article: not found
Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources.
- Record: found
- Abstract: found
- Article: not found