- Record: found
- Abstract: found
- Article: found
Genetic and epigenetic differentiation in response to genomic selection for avian lay date
Read this article at
Abstract
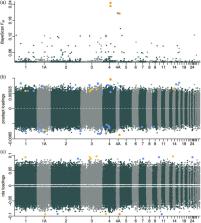
Anthropogenic climate change has led to globally increasing temperatures at an unprecedented pace and, to persist, wild species have to adapt to their changing world. We, however, often fail to derive reliable predictions of species' adaptive potential. Genomic selection represents a powerful tool to investigate the adaptive potential of a species, but constitutes a ‘blind process’ with regard to the underlying genomic architecture of the relevant phenotypes. Here, we used great tit ( Parus major) females from a genomic selection experiment for avian lay date to zoom into this blind process. We aimed to identify the genetic variants that responded to genomic selection and epigenetic variants that accompanied this response and, this way, might reflect heritable genetic variation at the epigenetic level. We applied whole genome bisulfite sequencing to blood samples of individual great tit females from the third generation of bidirectional genomic selection lines for early and late lay date. Genomic selection resulted in differences at both the genetic and epigenetic level. Genetic variants that showed signatures of selection were located within genes mostly linked to brain development and functioning, including LOC107203824 ( SOX3‐like). SOX3 is a transcription factor that is required for normal hypothalamo‐pituitary axis development and functioning, an essential part of the reproductive axis. As for epigenetic differentiation, the early selection line showed hypomethylation relative to the late selection line. Sites with differential DNA methylation were located in genes important for various biological processes, including gonadal functioning (e.g., MSTN and PIK3CB). Overall, genomic selection for avian lay date provided insights into where within the genome the heritable genetic variation for lay date, on which selection can operate, resides and indicates that some of this variation might be reflected by epigenetic variants.
Related collections
Most cited references125
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found
Fast gapped-read alignment with Bowtie 2.
- Record: found
- Abstract: not found
- Article: not found