- Record: found
- Abstract: found
- Article: not found
Natural products can be used in therapeutic management of COVID-19: Probable mechanistic insights
Read this article at
Abstract
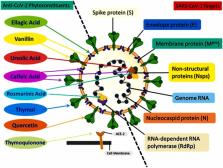
The unexpected emergence of the new Coronavirus disease (COVID-19) has affected more than three hundred million individuals and resulted in more than five million deaths worldwide. The ongoing pandemic has underscored the urgent need for effective preventive and therapeutic measures to develop anti-viral therapy. The natural compounds possess various pharmaceutical properties and are reported as effective anti-virals. The interest to develop an anti-viral drug against the novel severe acute respiratory syndrome Coronavirus (SARS-CoV-2) from natural compounds has increased globally. Here, we investigated the anti-viral potential of selected promising natural products. Sources of data for this paper are current literature published in the context of therapeutic uses of phytoconstituents and their mechanism of action published in various reputed peer-reviewed journals. An extensive literature survey was done and data were critically analyzed to get deeper insights into the mechanism of action of a few important phytoconstituents. The consumption of natural products such as thymoquinone, quercetin, caffeic acid, ursolic acid, ellagic acid, vanillin, thymol, and rosmarinic acid could improve our immune response and thus possesses excellent therapeutic potential. This review focuses on the anti-viral functions of various phytoconstituent and alkaloids and their potential therapeutic implications against SARS-CoV-2. Our comprehensive analysis provides mechanistic insights into phytoconstituents to restrain viral infection and provide a better solution through natural, therapeutically active agents.
Graphical Abstract
Related collections
Most cited references192
- Record: found
- Abstract: found
- Article: not found
A Novel Coronavirus from Patients with Pneumonia in China, 2019
- Record: found
- Abstract: found
- Article: not found
Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding

- Record: found
- Abstract: found
- Article: found

