- Record: found
- Abstract: found
- Article: found
Myosin-II mediated traction forces evoke localized Piezo1-dependent Ca 2+ flickers
Read this article at
Abstract
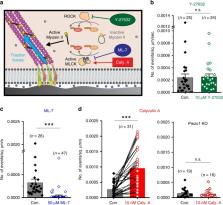
Piezo channels transduce mechanical stimuli into electrical and chemical signals to powerfully influence development, tissue homeostasis, and regeneration. Studies on Piezo1 have largely focused on transduction of “outside-in” mechanical forces, and its response to internal, cell-generated forces remains poorly understood. Here, using measurements of endogenous Piezo1 activity and traction forces in native cellular conditions, we show that cellular traction forces generate spatially-restricted Piezo1-mediated Ca 2+ flickers in the absence of externally-applied mechanical forces. Although Piezo1 channels diffuse readily in the plasma membrane and are widely distributed across the cell, their flicker activity is enriched near force-producing adhesions. The mechanical force that activates Piezo1 arises from Myosin II phosphorylation by Myosin Light Chain Kinase. We propose that Piezo1 Ca 2+ flickers allow spatial segregation of mechanotransduction events, and that mobility allows Piezo1 channels to explore a large number of mechanical microdomains and thus respond to a greater diversity of mechanical cues.
all
Ellefsen et al. monitor Piezo-dependent Calcium signals in live cells by TIRF and super-resolution microscopy and find that Ca 2+ flickers localize to areas of high traction force. They show that Myosin II activity and MLCK are needed for the generation of Piezo Ca 2+ signals and that Piezo1 channels are mobile in the plasma membrane.
Related collections
Most cited references50
- Record: found
- Abstract: found
- Article: not found
Piezos are pore-forming subunits of mechanically activated channels
- Record: found
- Abstract: found
- Article: not found
Piezo1, a mechanically activated ion channel, is required for vascular development in mice.
- Record: found
- Abstract: found
- Article: not found