- Record: found
- Abstract: found
- Article: not found
Nanosecond Time Scale Motions in Proteins Revealed by High-Resolution NMR Relaxometry
Read this article at
Abstract
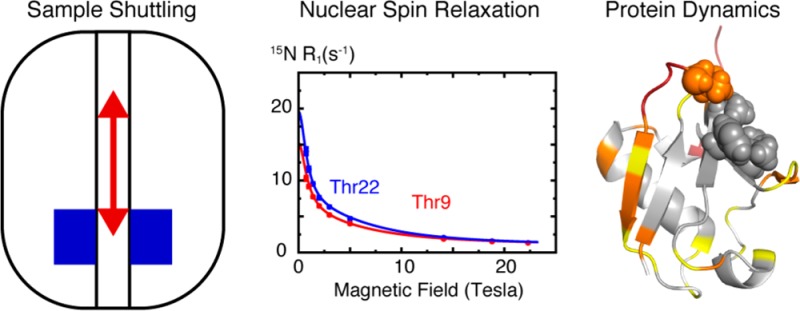
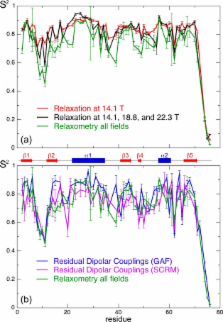
Understanding the molecular determinants underlying protein function requires the characterization of both structure and dynamics at atomic resolution. Nuclear relaxation rates allow a precise characterization of protein dynamics at the Larmor frequencies of spins. This usually limits the sampling of motions to a narrow range of frequencies corresponding to high magnetic fields. At lower fields one cannot achieve sufficient sensitivity and resolution in NMR. Here, we use a fast shuttle device where the polarization builds up and the signals are detected at high field, while longitudinal relaxation takes place at low fields 0.5 < B 0 < 14.1 T. The sample is propelled over a distance up to 50 cm by a blowgun-like system in about 50 ms. The analysis of nitrogen-15 relaxation in the protein ubiquitin over such a wide range of magnetic fields offers unprecedented insights into molecular dynamics. Some key regions of the protein feature structural fluctuations on nanosecond time scales, which have so far been overlooked in high-field relaxation studies. Nanosecond motions in proteins may have been underestimated by traditional high-field approaches, and slower supra-τ c motions that have no effect on relaxation may have been overestimated. High-resolution relaxometry thus opens the way to a quantitative characterization of nanosecond motions in proteins.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: found
Rapid planetesimal formation in turbulent circumstellar discs
- Record: found
- Abstract: found
- Article: not found
Ubiquitin-binding domains - from structures to functions.
- Record: found
- Abstract: not found
- Article: not found
