- Record: found
- Abstract: found
- Article: found
mGOASVM: Multi-label protein subcellular localization based on gene ontology and support vector machines
Read this article at
Abstract
Background
Although many computational methods have been developed to predict protein subcellular localization, most of the methods are limited to the prediction of single-location proteins. Multi-location proteins are either not considered or assumed not existing. However, proteins with multiple locations are particularly interesting because they may have special biological functions, which are essential to both basic research and drug discovery.
Results
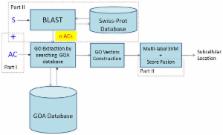
This paper proposes an efficient multi-label predictor, namely mGOASVM, for predicting the subcellular localization of multi-location proteins. Given a protein, the accession numbers of its homologs are obtained via BLAST search. Then, the original accession number and the homologous accession numbers of the protein are used as keys to search against the Gene Ontology (GO) annotation database to obtain a set of GO terms. Given a set of training proteins, a set of T relevant GO terms is obtained by finding all of the GO terms in the GO annotation database that are relevant to the training proteins. These relevant GO terms then form the basis of a T-dimensional Euclidean space on which the GO vectors lie. A support vector machine (SVM) classifier with a new decision scheme is proposed to classify the multi-label GO vectors. The mGOASVM predictor has the following advantages: (1) it uses the frequency of occurrences of GO terms for feature representation; (2) it selects the relevant GO subspace which can substantially speed up the prediction without compromising performance; and (3) it adopts an efficient multi-label SVM classifier which significantly outperforms other predictors. Briefly, on two recently published virus and plant datasets, mGOASVM achieves an actual accuracy of 88.9% and 87.4%, respectively, which are significantly higher than those achieved by the state-of-the-art predictors such as iLoc-Virus (74.8%) and iLoc-Plant (68.1%).
Conclusions
mGOASVM can efficiently predict the subcellular locations of multi-label proteins. The mGOASVM predictor is available online at http://bioinfo.eie.polyu.edu.hk/mGoaSvmServer/mGOASVM.html.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences.
- Record: found
- Abstract: found
- Article: not found
Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation.
- Record: found
- Abstract: not found
- Article: not found