- Record: found
- Abstract: found
- Article: found
Disentangling the role of poultry farms and wild birds in the spread of highly pathogenic avian influenza virus in Europe
Read this article at
Abstract
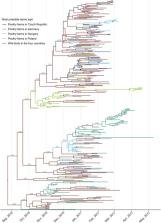
In winter 2016–7, Europe was severely hit by an unprecedented epidemic of highly pathogenic avian influenza viruses (HPAIVs), causing a significant impact on animal health, wildlife conservation, and livestock economic sustainability. By applying phylodynamic tools to virus sequences collected during the epidemic, we investigated when the first infections occurred, how many infections were unreported, which factors influenced virus spread, and how many spillover events occurred. HPAIV was likely introduced into poultry farms during the autumn, in line with the timing of wild birds’ migration. In Germany, Hungary, and Poland, the epidemic was dominated by farm-to-farm transmission, showing that understanding of how farms are connected would greatly help control efforts. In the Czech Republic, the epidemic was dominated by wild bird-to-farm transmission, implying that more sustainable prevention strategies should be developed to reduce HPAIV exposure from wild birds. Inferred transmission parameters will be useful to parameterize predictive models of HPAIV spread. None of the predictors related to live poultry trade, poultry census, and geographic proximity were identified as supportive predictors of HPAIV spread between farms across borders. These results are crucial to better understand HPAIV transmission dynamics at the domestic–wildlife interface with the view to reduce the impact of future epidemics.
Related collections
Most cited references54

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability

- Record: found
- Abstract: found
- Article: found
Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7
- Record: found
- Abstract: not found
- Article: not found