- Record: found
- Abstract: found
- Article: found
Spatial and Single-Cell Transcriptomics Reveal a Cancer-Associated Fibroblast Subset in HNSCC That Restricts Infiltration and Antitumor Activity of CD8 + T Cells
Read this article at
Abstract
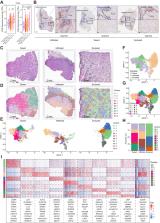
Spatial analysis identifies IFN-induced MHC-I hiGal9 + CAFs that form a trap for CD8 + T cells, providing insights into the complex networks in the tumor microenvironment that regulate T-cell infiltration and function.
Abstract
Although immunotherapy can prolong survival in some patients with head and neck squamous cell carcinoma (HNSCC), the response rate remains low. Clarification of the critical mechanisms regulating CD8 + T-cell infiltration and dysfunction in the tumor microenvironment could help maximize the benefit of immunotherapy for treating HNSCC. Here, we performed spatial transcriptomic analysis of HNSCC specimens with differing immune infiltration and single-cell RNA sequencing of five pairs of tumor and adjacent tissues, revealing specific cancer-associated fibroblast (CAF) subsets related to CD8 + T-cell infiltration restriction and dysfunction. These CAFs exhibited high expression of CXCLs (CXCL9, CXCL10, and CXCL12) and MHC-I and enrichment of galectin-9 (Gal9). The proportion of MHC-I hiGal9 + CAFs was inversely correlated with abundance of a TCF1 +GZMK + subset of CD8 + T cells. Gal9 on CAFs induced CD8 + T-cell dysfunction and decreased the proportion of tumor-infiltrating TCF1 +CD8 + T cells. Collectively, the identification of MHC-I hiGal9 + CAFs advances the understanding of the precise role of CAFs in cancer immune evasion and paves the way for more effective immunotherapy for HNSCC.
Related collections
Most cited references39

- Record: found
- Abstract: found
- Article: found
GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses
- Record: found
- Abstract: found
- Article: not found
TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells.

- Record: found
- Abstract: found
- Article: found